PBSP (Phosphate Binding Site Predictor) is a stand-alone command line program that predicts phosphate binding sites from protein structures. The approach leverages a modified energy-based ligand binding sites identification method and combines it with reverse focused docking in which a phosphate probe is used to improve the accuracy and selectivity of predictions. Here, we developed two PBSP models, PBSP_R by rigid docking and PBSP_F by flexible docking.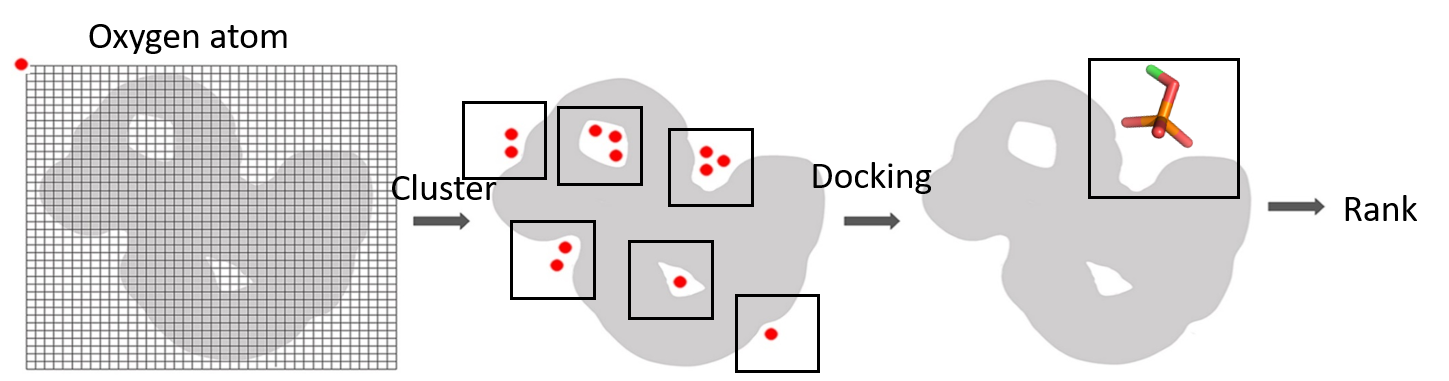
Download PBSP and Benchmark Datasets
(1) PBSP_R (run on linux 64 bit)
(2) PBSP_F (run on linux 64 bit)
(3) Bound dataset
a non-redundant bound dataset of 97 phosphate binding proteins containing 106 phosphate binding sites (proteins < 50% sequence identity).
(4) Unbound dataset
a non-redundant unbound dataset of 63 phosphate binding proteins containing 67 phosphate binding sites (proteins < 50% sequence identity).
Installation
PBSP relies on and contains third-party softwares called AutoDockFR and AutoSite. The PBSP executable should be place in the same folder as the modified ADFRsuite. A linux system(64 bit) are required to run the scripts that are provided to automate the calculations. The suite does not require any special installation procedure. The archive downloaded from the web must be uncompressed and can then be placed anywhere.
Usage
The PBSP executable provided in the root directory of the suite allows the user to carry out phosphate binding site identification using focused reverse docking. An example of the most basic use:
PBSP_R --input=protein.pdb --probe=CH3_PO4.pdb --outdir=/lustre/home/zclu/
PBSP_R -i protein.pdb -p CH3_PO4.pdb -o /lustre/home/zclu/
CH3PO42- is a phosphate probe which is docked to the potential phosphate binding pocket to generate phosphate binding modes.
Tips
1) PBSP_R is a executable which are generated by from the source Python script 'PBSP_R.py' by PyInstaller.
2) PyInstaller is a program that converts Python programs into stand-alone executables, under Windows, Linux, and Mac OS X. PyInstaller can bundles a Python application and all its dependencies into a single package. The user can run the packaged app without installing a Python interpreter or any modules.
3) If you are familiar with Python programming languages, you can use and modify the the script 'PBSP_R.py'. The 'PBSP_R.py' script is written in Python 3, and is placed in the script floder.
Outputs
(1) Predicted phosphate binding sites
Output file: 1d4w_C_281_predicted_phosphate_binding_sites.txt
Rank Pocket Predicted Phosphate Binding Sites Predicted Binding Energy
1 1d4w_C_281_pocket000 ARG_55,GLU_35,SER_36,SER_34,CYS_42,ARG_32 -5.7
2 1d4w_C_281_pocket001 LYS_81,LYS_79 -5.5
3 1d4w_C_281_pocket013 ARG_75,LYS_89,GLN_92 -3.7
4 1d4w_C_281_pocket008 LYS_74,THR_68 -3.5
5 1d4w_C_281_pocket003 SER_65,LYS_74,GLU_67,SER_57 -3.4
6 1d4w_C_281_pocket009 SER_65,LYS_74,SER_57 -3.4
7 1d4w_C_281_pocket002 GLY_93,ASP_91,PRO_97,ILE_94,GLN_92 -3.3
8 1d4w_C_281_pocket021 ASN_82,ARG_78 -3.2
9 1d4w_C_281_pocket022 TYR_41,MET_1 -3.1
10 1d4w_C_281_pocket004 ASP_2,LYS_104,GLY_9 -2.9
11 1d4w_C_281_pocket010 GLN_58,SER_57 -1.7
12 1d4w_C_281_pocket011 GLU_14,ARG_13,GLU_35 -1.7
13 1d4w_C_281_pocket006 GLY_39,PRO_38,VAL_40,SER_34,ASP_33 -1.6
14 1d4w_C_281_pocket012 TYR_29,GLN_88 -1.6
15 1d4w_C_281_pocket015 ARG_32,ARG_13,SER_12,GLU_35 -1.1
16 1d4w_C_281_pocket024 GLN_99,TYR_100 -1.1
17 1d4w_C_281_pocket023 GLU_103,VAL_102 -1.0
(2) Predicted binding modes
Output file: 1d4w_C_281_pocket000_complex.pdb
Citation
PBSP relies on and contains third-party softwares called AutoDockFR and AutoSite. if you used PBSP in your work, please cite:
1) Z.C. Lu, F. Jiang and Y.D. Wu, Phosphate binding sites prediction in phosphorylation-dependent protein-protein interactions, in revision
2) P.A. Ravindranath, S. Forli, D.S. Goodsell, A.J. Olson and M.F. Sanner, AutoDockFR: Advances in Protein-Ligand Docking with Explicitly Specified Binding Site Flexibility, PLoS Comput Biol (2015). DOI:10.1371/journal.pcbi.1004586
3) P.A. Ravindranath and M.F. Sanner, AutoSite: an automated approach for pseudo- ligands prediction - From ligand binding sites identification to predicting key ligand atoms, Bioinformatics (2016). DOI:10.1093/bioinformatics/btw367
Please contact Zheng-Chang Lu (luzc@pku.edu.cn) if you have an idea for a project proposal or any questions.